Bacteria like to make their gooey homes along boundary surfaces to protect themselves from inclement conditions and attacks. So far, however, the architectural strategy of these bacteria has eluded researchers. A team working with the scientists Anne Diehl (Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP)) and Yvette Roske (Max Delbrück Center for Molecular Medicine (MDC)) watched Bacillus subtilis build shelters and reported their observations in PNAS. Surprisingly, bacteria form a precursor of the major building block for the biofilm (the protein TasA) inside their cells. Once outside the bacteria, these building blocks form moderately long chains known as fibrils. Like a matrix, these fibrils serve to stabilize the frame house.
Biofilms are a potential hazard for people because inside these protective shelters, bacteria are safe from attack by the immune system or antibiotics. To hinder the formation of biofilms and guarantee the efficacy of antibiotics, scientists need to know the structure of the building blocks.The TasA protein surprises with dynamic changeThe scientists at the FMP and MDC seized upon a suggestion made by the B. subtilis expert Kürşad Turgay at the University Hannover (Germany) and revealed the molecular structure of TasA as the dominant biofilm protein.
Anne Diehl (FMP Department of NMR-Assisted Structural Research under Hartmut Oschkinat) started the synthesis of TasA and studied the various structural forms of the protein. She noticed how fast TasA changes its structure. Her work hit an unexpected snag: "In my 32 years of elucidating protein structures, I never worked with a protein as dynamic as this one. Even after a short time, the soluble TasA proteins self-assemble into a gel-like consistency", reported Anne Diehl looking back at the successful characterization of the individual protein structures.
A robust core with flexible loops
Yvette Roske (MDC Department of Macromolecular Structures and Interactions under Udo Heinemann) found a conceivable explanation for the behavior of the TasA protein. Starting with freshly purified TasA, she grew single crystals and analyzed them using the high-energy X-rays of the synchrotron facility BESSY in Berlin-Adlershof. This allowed her to map out the folded three-dimensional structure of the TasA protein. "As it turned out, large regions of the TasA structure are highly organized. The large portion of ß-pleated sheet elements gives the protein the robust core. This core seems decorated with flexible loops", summarizes Yvette Roske. Interestingly, the amino acid arginine is not present in TasA. On average, the basic amino acid arginine accounts for 10 % of the amino acids in a protein. This amounts to double that expected if all 20 amino acids would occur in equal frequency.
Anne Diehl elaborates, "There must be a reason why this protein completely excludes one common amino acid."Frequently, arginine marks the target point for proteases (housekeeping proteins, which cut the peptide bonds of other proteins). The absence of arginine could feasibly explain the extraordinary stability of TasA against proteases, making it a strong buttress for the biofilm shelters.
TasA presence may also explain why Bacillus subtilis is not pathogenic
The amino acid sequence of TasA actually resembles the sequence of a protease named Camelysin. Many pathogenic bacilli use Camelysin instead of TasA for their biofilms. Based on their TasA structure, the scientists were able to construct a model for Camelysin. "Our structural model for Camelysin suggests that the three-dimensional folding of Camelysin resembles the folding of TasA", Yvette Roske points out. The Camelysin of the anthrax strain B. anthracis is a protease. In contrast, the TasA of the harmless B. subtilis lacks this activity.
In the course of evolution, TasA seems to have lost its protease activity and with it its pathogenic properties. Research into biofilms has now entered its next phase. The FMP and MDC teams are already pursuing new avenues of research. Using solid state NMR, the teams discovered the re-orientation of unorganized flexible TasA sections when the TasA building blocks assemble into fibrils. Further studies of the fibrils will help scientists to reveal the secret of biofilm stability. The next results may offer new approaches to fighting pathogens.
Sources
Anne Diehl, Yvette Roske, Linda Ball, Anup Chowdhury, Matthias Hiller, Noel Molière, Regina Kramer, Daniel Stöppler, Catherine L. Worth, Brigitte Schlegel, Martina Leidert, Nils Cremer, Natalja Erdmann, Daniel Lopez, Heike Stephanowitz, Eberhard Kraus, Barth-Jan van Rossum, Peter Schmieder, Udo Heinemann, Kürşad Turgay, Ümit Akbey and Hartmut Oschkinat. Structural changes of TasA in biofilm formation of Bacillus subtilis, PNAS 12. March 2018, DOI 10.1073/pnas.1718102115
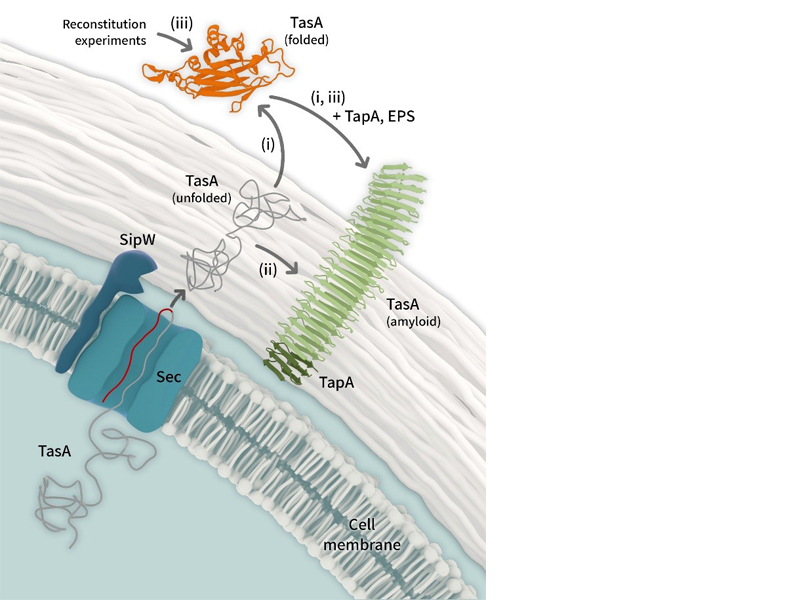
Processes leading to Bacillus subtilis fibre formation after TasA
secretion (i,ii) and from externally provided protein in reconstitution
experiments (iii) with tasA mutants. In all cases, but in particular for
pathway (iii), a supporting role of TapA or Exopolysaccharides (not
shown) is expected. Visualization: Barth van Rossum, FMP
Contact
| Professor Dr. Hartmut Oschkinat Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP) oschkinatfmp-berlin.de Telephone: 49 (0) 309 479 3160 leibniz-fmp.de/oschkinat Dr. Anne Diehl Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP) diehlfmp-berlin.de Telephone: 49 (0) 309 479 3310 Public Relations Silke Oßwald Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP) Telephone: +49 (0) 309 479 3104 Email: osswaldfmp-berlin.de |